An AAPM Grand Challenge
Overview
The American Association of Physicists in Medicine (AAPM) is sponsoring the Quantitative Intravoxel Incoherent Motion (IVIM) Diffusion MRI (dMRI) Reconstruction Challenge, leading up to the 2024 AAPM Annual Meeting & Exhibition. We invite participants to develop image reconstruction and model fitting methods to improve the accuracy and robustness of quantitative parameter estimation for the widely used IVIM model of dMRI [1]. Both deep learning (DL) and non-DL approaches are welcome. Methods can operate in the image-domain, in the k-space domain, or in a combination of both. In this Challenge, participants will be provided with k-space data of breast dMRI generated via rigorous simulations that accurately represent the dMRI signal generation process associated with the IVIM model across a range of diffusion weighting (b-values). The participants will be asked to derive IVIM parameter maps and compete for the most accurate reconstruction results. The two top-performing teams (only one member per team) will be awarded complimentary meeting registration to present on their methodologies during the 2024 AAPM Annual Meeting & Exhibition in Los Angeles, CA from July 21-25, 2024 (in-person attendance is required). The challenge organizers will summarize the challenge results in a journal publication after the Annual Meeting.
Background
Diffusion MRI (dMRI) has been extensively employed over the years for the diagnosis of diseases and is increasingly used to guide radiation therapy and assess treatment responses. dMRI captures the random motion of water protons influenced by tissue microstructure, thus offering valuable insights into clinically significant tissue microstructural properties [2, 3]. Unlike conventional MRI reconstruction problems, which focus on retrieving anatomical images from measured k-space data, dMRI reconstruction aims to quantitatively determine images of biophysical tissue microstructural parameters. However, the estimation of these parameters is often accompanied by considerable uncertainties due to the complex inverse problem posed by the highly nonlinear nature of dMRI signal models [4, 5], particularly in scenarios with low signal-to-noise ratios (SNRs) resulting from fast image acquisition and physiological motion. The substantial variation and bias in parameter estimation hinder the interpretation of results and impede the reliable clinical application of dMRI in tissue characterization and longitudinal evaluations.
Objective
The proposed IVIM-dMRI Reconstruction Challenge aims to enhance accuracy and robustness of quantitative dMRI reconstruction, with a focus on the widely used and clinically significant IVIM model [1]. The IVIM model enables simultaneous assessment of perfusion and diffusion by fitting the dMRI signal to a biexponential model that captures both water molecular diffusion and blood microcirculation. The primary task of this Challenge is to achieve quantitative reconstruction of IVIM-dMRI tissue parametric maps, specifically fractional perfusion (![]() ) related to microcirculation, pseudo-diffusion coefficient (
) related to microcirculation, pseudo-diffusion coefficient (![]() ), and true diffusion coefficient (
), and true diffusion coefficient (![]() ), from the provided k-space data, and strive for the most accurate reconstruction results.
), from the provided k-space data, and strive for the most accurate reconstruction results.
IVIM-dMRI Model and Challenge Data
Details of the data generation methodology will be provided on the IVIM-dMRI Reconstruction Challenge website.
In IVIM-dMRI, a series of MR images are acquired, each under a diffusion weight ![]() . The MRI signal of each voxel
. The MRI signal of each voxel ![]() follows a bi-exponential equation as
follows a bi-exponential equation as ![]() , where
, where ![]() is signal intensity at
is signal intensity at ![]() = 0 s/mm2 For each diffusion weight, the measured complex k-space data
= 0 s/mm2 For each diffusion weight, the measured complex k-space data ![]() is related to
is related to ![]() via the standard Fourier Transform (FT) procedure with noise as
via the standard Fourier Transform (FT) procedure with noise as
![]()
where ![]() denotes FT operation, and
denotes FT operation, and ![]() is an independent and identically distributed (i.i.d.) Gaussian noise.
is an independent and identically distributed (i.i.d.) Gaussian noise.
 In this challenge, participants are given k-space data
In this challenge, participants are given k-space data ![]() at a series of known
at a series of known ![]() values, and are tasked to reconstruct images of
values, and are tasked to reconstruct images of ![]() ,
, ![]() , and
, and ![]() .
.
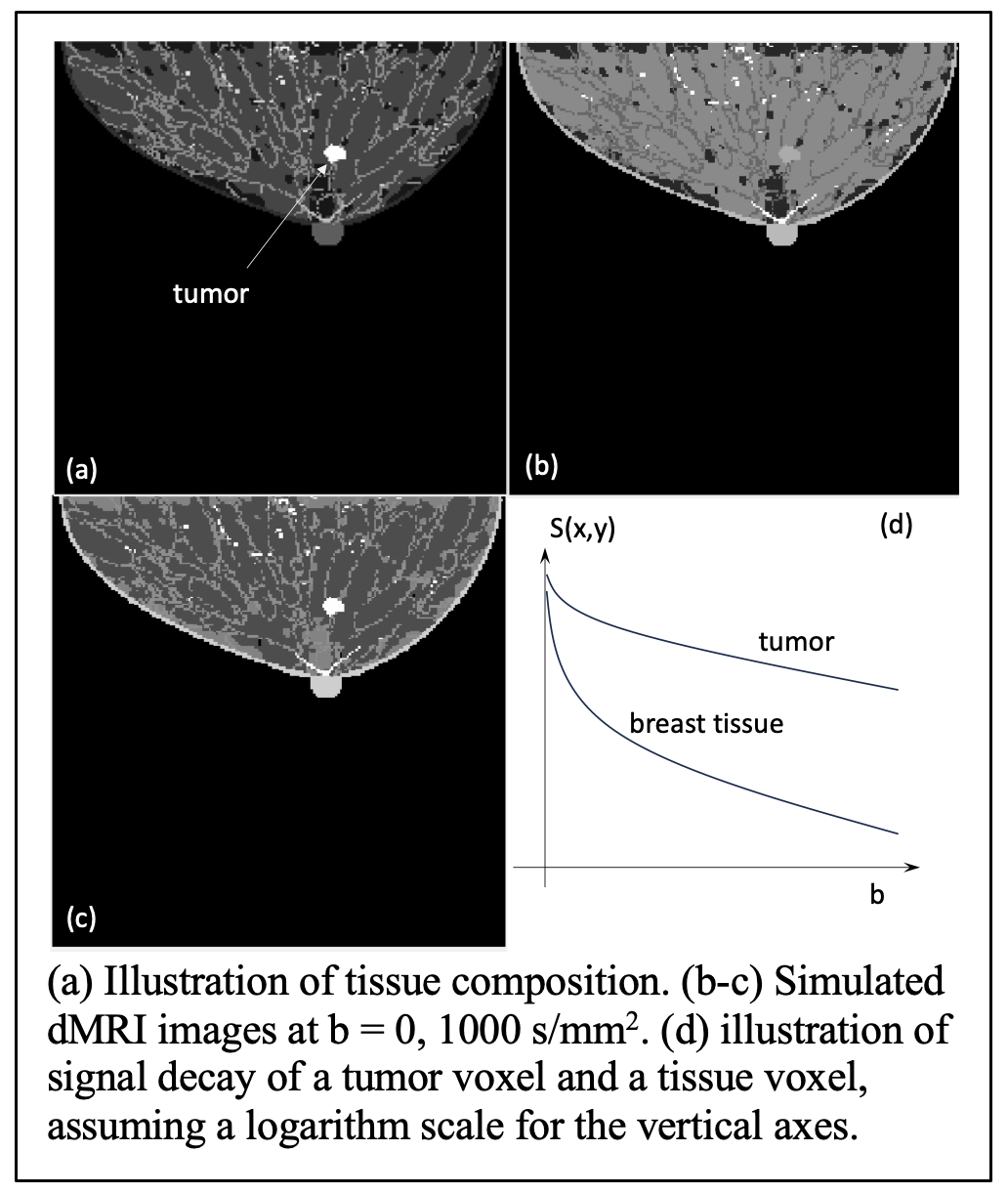
We generated simulated breast MR images using the VICTRE breast phantom. These breast phantom images will depict realistic breast anatomy consisting of various types of normal breast tissues and tumor tissue types demonstrating intratumoral he terogeneity (See the figure). In this figure, (a) depicts tissue compositions with each voxel value being an integer label for illustration purpose.
For each tissue type, we assigned known values for ![]() ,
, ![]() and
and ![]() serving as the gold standard. The values for
serving as the gold standard. The values for ![]() ,
, ![]() and
and ![]() will adhere to the parameters established in prior scientific literature, ensuring their alignment with realistic biological interpretations. Based on the tissue specific MRI properties, we generated the images
will adhere to the parameters established in prior scientific literature, ensuring their alignment with realistic biological interpretations. Based on the tissue specific MRI properties, we generated the images ![]() . Figure (b-c) shows images at
. Figure (b-c) shows images at ![]() = 0 and 1000 s/mm2 in the absence of noise. (d) illustrates the signal decay for a tumor and a tissue voxel.
= 0 and 1000 s/mm2 in the absence of noise. (d) illustrates the signal decay for a tumor and a tissue voxel.
Finally, based on the simulated ![]() at a series of
at a series of ![]() values, FT was performed for image at each
values, FT was performed for image at each ![]() value and then noise was added, yielding the k-space data available for the reconstruction.
value and then noise was added, yielding the k-space data available for the reconstruction.
How the Challenge Works
This challenge consists of three phases:
Phase I (training and development phase)
Participants will be given access to the codes with a description of the simulation process for generating k-space datasets with a range of b-values from the known tissue microstructural parametric maps. A training dataset including 1000 cases will be provided for the participants to develop reconstruction models. Each case will include
- A series of images with 200 × 200 pixels to represent known tissue microstructural parametric maps.
- An image with 200 × 200 pixels representing known tissue types of each voxel.
- Ground truth noiseless
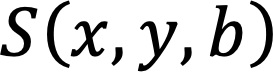 images with 200 × 200 pixels at a series of
images with 200 × 200 pixels at a series of 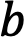 values.
values. - Simulated noisy k-space data of complex images with 200 × 200 pixels at corresponding
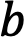 values.
values.
To facilitate the understanding of the dMRI model and our data, we will provide a Python script that performs inverse FT to recover ![]() from k-space data and then derives tissue parameter maps via pixel-wise data fitting. The script will also include codes to write the results in a specific format for automatic evaluation by the Challenge webpage.
from k-space data and then derives tissue parameter maps via pixel-wise data fitting. The script will also include codes to write the results in a specific format for automatic evaluation by the Challenge webpage.
Phase II (validation and refinement phase)
Participants will validate their algorithms using provided validation dataset and submit their reconstruction results through the Challenge webpage. The validation dataset will consist of 10 cases, each including noisy k-space data of complex images with 200 × 200 pixels at corresponding ![]() values. Ground truth of the validation datasets will not be provided.
values. Ground truth of the validation datasets will not be provided.
After the participants submit their results to the Challenge webpage, the results will be evaluated using predetermined evaluation metrics, and a leaderboard will display the performance of different participants. At this phase, the number of submissions is unlimited.
- Required submission:
- Reconstructed tissue microstructure parametric maps for the validation cases.
Phase III (testing and final scoring phase)
Participants will run their algorithms on provided test dataset and submit their reconstruction results through the Challenge website. The test dataset will consist of 100 cases, each including noisy k-space data of complex images with 200 × 200 pixels at corresponding ![]() values. Ground truth of the test datasets will not be provided.
values. Ground truth of the test datasets will not be provided.
After the participants submit their results to the Challenge webpage, the results will be evaluated using predetermined evaluation metrics, and a leaderboard will display the performance of different participants. At this phase, each participant team is allowed a maximum of three submissions. Required submission:
- Reconstructed tissue microstructure parametric maps for the test cases.
- A one-page technical note describing the reconstruction algorithm.
Evaluation Metrics
The accuracy of the reconstructed IVIM-dMRI parameters will be evaluated using the following metrics (see detailed information in at the Challenge website)
- Primary metric: relative Root-mean-square-error (rRMSE) for each of the three IVIM parameters will be computed, averaging the results over the provided validation/testing dataset.
- Secondary metrics: We will compute the rRMSE for tumor area to investigate performance of the reconstruction algorithms in clinically relevant regions. If needed, the secondary metrics averaged over the four regions will be used to break ties under the primary evaluation metric.
Get Started
- Register to get access via the Challenge website
- Get access to the codes and training data after approval
- Develop your reconstruction algorithm
- Download the validation dataset and submit your results to receive preliminary scores
- Download the test dataset and submit your final results
Important Dates
- January 15, 2024: Phase I starts. Registration opens. Training dataset and script are made available.
- February 15, 2024: Phase II starts. Validation datasets are made available. Participants can submit preliminary results and receive feedback on relative scoring for unlimited number of times.
- May 15, 2024: Phase III starts. Final test datasets are made available.
- June 3, 2024: Deadline for the final submission of results and a one-page summary describing the algorithm (midnight, ET)
- June 10, 2024: Participants are notified with challenge results and winners (top 2) are announced.
- July 21-25, 2024: AAPM Annual Meeting & Exhibition: top two teams will present on their work during a dedicated challenge session.
- September 2024: The challenge organizers summarize the grand challenge in a journal paper. Datasets and scoring routines will be made public.
Results, Prizes and Publication Plan
At the conclusion of the challenge, the following information will be provided to each participant:
- The evaluation results for the submitted cases
- The overall ranking among the participants
The top 2 participants (one member from each team only):
- Will present their algorithm and results at the AAPM Annual Meeting & Exhibition (July 21-25, 2024, Los Angeles, CA). In-person attendance is required.
- Will be awarded complimentary registration to the AAPM Annual Meeting & Exhibition.
A manuscript summarizing the challenge results will be submitted for publication after the AAPM Annual Meeting & Exhibition.
Terms and Conditions
The following rules apply to those who register and download the data:
- Anonymous participation is not allowed.
- Only one representative can register per team.
- Members from the host institution and the organizers’ groups cannot participate. (Please review AAPM's participant COI statement.)
- Entry by commercial entities is permitted but should be disclosed; conflict of interest attestations will be required for all participants upon registration.
- Once participants submit their results to the IVIM-dMRI Reconstruction challenge, they will be considered fully vested in the challenge, so that their performance results will become part of any presentations, publications, or subsequent analyses derived from the Challenge at the discretion of the organizers.
- Participants summarize their algorithms in a document to submit at the end of Phase II.
- The downloaded code, datasets or any data derived from these datasets, may not be redistributed under any circumstances to persons not belonging to the registered teams.
- Data and code downloaded from this site may only be used for the purpose of scientific studies and may not be used for commercial purposes. Acknowledge the source of data by:
- C. Graff, “A new, open-source, multi-modality digital breast phantomExternal Link Disclaimer”, Proc. SPIE 9783, Medical Imaging 2016: Physics of Medical Imaging, 978309.
- A. Badano, C. G. Graff, A. Badal, D. Sharma, R. Zeng, F. W. Samuelson, S. Glick, and K. J. Myers, "Evaluation of Digital Breast Tomosynthesis as Replacement of Full-Field Digital Mammography Using an In Silico Imaging TrialExternal Link Disclaimer", JAMA Network Open. 2018; 1(7).
- <TBD paper to be submitted by the challenge organizers>
Organizers
- Xun Jia, Ph.D., DABR, FAAPM (Lead Organizer) (Johns Hopkins University School of Medicine)
- Jie Deng, Ph.D., DABMP (University of Texas Southwestern Medical Center)
- Junghoon Lee, Ph.D. (Johns Hopkins University School of Medicine)
- Ahad Ollah Ezzati, Ph.D. (Johns Hopkins University School of Medicine)
- Yan Dai (University of Texas Southwestern Medical Center)
- Xiaoyu Hu, Ph.D. (Johns Hopkins University School of Medicine)
- The AAPM Working Group on Grand Challenges
Contacts
For further information, please contact the lead organizer, Xun Jia (xunjia@jhu.edu) or AAPM staff member, Emily Townley (emily@aapm.org).
References
- Le Bihan, D., et al., MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology, 1986. 161(2): p. 401-407.
- Koh, D.-M. and D.J. Collins, Diffusion-weighted MRI in the body: applications and challenges in oncology. American Journal of Roentgenology, 2007. 188(6): p. 1622-1635.
- Iima, M. and D. Le Bihan, Clinical intravoxel incoherent motion and diffusion MR imaging: past, present, and future. Radiology, 2016. 278(1): p. 13-32.
- Barbieri, S., et al., Impact of the calculation algorithm on biexponential fitting of diffusion-weighted MRI in upper abdominal organs. Magnetic resonance in medicine, 2016. 75(5): p. 2175-2184.
- Landaw, E. and J. DiStefano 3rd, Multiexponential, multicompartmental, and noncompartmental modeling. II. Data analysis and statistical considerations. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology, 1984. 246(5): p. R665-R677.



















