An AAPM Grand Challenge
Overview:
The American Association of Physicists in Medicine (AAPM) is sponsoring the Generalizable Dose Prediction for Heterogenous Multi-Cohort and Multi-Site Radiotherapy Planning (GDP-HMM) Challenge, leading up to the 2025 AAPM Annual Meeting & Exhibition. We invite participants to develop artificial intelligence models to contribute to generalizable 3D dose prediction for radiotherapy (RT). In this Challenge, participants will be provided with CT, PTV & OAR masks, beam geometries, and other meta information as the input, and reference RT dose as training ground truth which is generated by Varian Eclipse treatment system. The participants will be asked to develop deep learning models to predict dose distribution given patient data and beam setup but without planning optimization. The two top-performing teams (one member per team) will be awarded complimentary meeting registration to present on their methodologies during the AAPM Annual Meeting & Exhibition in Washington, DC from July 27-31, 2025 (in-person attendance is required). The Challenge organizers will summarize the Challenge results in a journal publication after the Annual Meeting.
Background:
Radiotherapy treatment planning is a complex task that involves designing an optimal plan based on a patient's anatomy and physician directives. This process is time-consuming and resource-intensive, with the quality of the plan heavily reliant on the planner's expertise. 3D artificial intelligence-based models for dose map prediction have been recently developed to facilitate the planning process for different cancers, such as head-and-neck [1], and lung [2]. However, most studies are conducted on institution-specific datasets and evaluation metrics. OpenKBP [3] has established the first dose prediction challenge in the domain in 2020, addressing these issues five years ago. We propose a more comprehensive Challenge that allows researchers to utilize state-of-the-art AI models while also addressing shortcomings of the previous OpenKBP Challenge with following:
- Assessing the applicability of dose prediction models in the downstream task of treatment planning optimization. We have used an optimization engine, Varian Eclipse System, to convert the predicted dose into a final deliverable plan. Additionally, we employ a set of clinical scorecards to evaluate the quality of the final plan.
- Evaluating the generalizability of the dose prediction models across various disease sites and cohorts from different institutions.
Objective:
The GDP-HMM Challenge aims to enhance the quality and consistency of radiotherapy treatment plans by advancing the current dose prediction research in the field. It focuses on evaluating the applicability and generalizability of these techniques while promoting reproducibility and transparency using a uniform benchmark dataset and evaluation criteria.
Challenge Data:
Details of the data generation methodology and download link will be provided on the GDP-HMM Challenge website.
We used the below pipeline to generate high quality RT plan data.
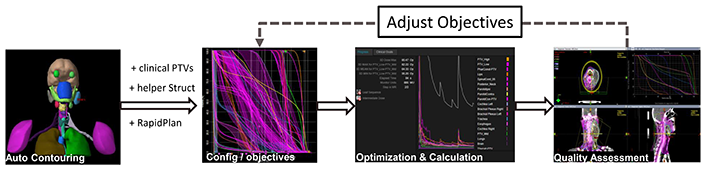
We use the CT and PTV masks from public datasets (cohorts will be disclosed after the Challenge), and create helper structures following the RapidPlan and ScoreCard in Varian Medical Affairs website https://medicalaffairs.varian.com/halcyon-case-studies. As shown in Figure 1, all the processes are run automatically.
How the Challenge Works:
This Challenge consists of three phases:
Phase I (training and development phase)
Participants will be given access to training data, data documentation, and code repository. The code repository includes data visualization, preprocess pipeline, baseline, evaluation code. Each RT plan includes:
- CT image, stored with Numpy format.
- PTV & OAR masks, and helper structures, stored with Numpy format.
- Angle list and generated angle/beam plates inspired by [2].
- Meta information including planning mode (IMRT or VMAT), treatment site (Head-and-Neck or Lung). cohort index, prescribed dose numbers.
- 3D dose distribution, stored with Numpy format.
With the provided code repository, participants can understand the data with visualization and description. We provide preprocess code and descriptions so the participant can either directly use the processed data or adjust the code to make custom changes. The PyTorch baseline provides data loader, network structure, loss functions to train the model, and the evaluation code can calculate the metric (Metric 1) based on the prediction and reference. The repository also includes the script to write the results in a specific format for automatic evaluation by the Challenge platform.
Phase II (validation and refinement phase)
The validation set has no overlap with the training set, and input data are provided but no ground truth (i.e., 3D dose distribution). The participant can their results to the Challenge platform. The results will be evaluated using predetermined evaluation metrics, and a leaderboard will display the performance of different participants. At this phase, the number of submissions is unlimited. The Metric 1 will be displayed real time. However, the Metric 2 will only be provided at three time points.
Required submission:
- predicted 3D dose distribution for each plan.
Phase III (testing and final scoring phase)
No testing data will be provided to participants. Participants need to make a docker for their algorithm. The results will be evaluated using predetermined evaluation metrics, and a leaderboard will display the performance of different participants. At this phase, each participant team is allowed a maximum of three submissions.
Required submission:
- Inference code in Docker.
- Reproducible training code.
- 1–2 pages technique report.
Note: the team has missing item(s) in the testing phase will not be considered for award.
Evaluation Metrics:
Metric 1: Mean Absolution Error (masked by body). The evaluation code will be provided to participants.
Metric 2: Plan quality scorecard when using the predicted dose for re-planning with Varian Eclipse. The documentation scorecard and evaluation methods are disclosed to participants, while this metric is calculated by organizers in both Phase II and III.
Get Started:
- Register to get access via the Challenge platform.
- Access to https://github.com/RiqiangGao/GDP-HMM_AAPMChallenge - retrieve data, read documentation, and run baseline.
- Develop your own AI algorithm.
- Download the validation dataset and submit your results to receive preliminary scores.
- Submit your final algorithm via Docker during test phase.
Important Dates:
- January 5, 2025: Phase I starts. Registration opens. Training dataset and script are made available.
- February 15, 2025: Phase II starts. Validation datasets are made available. Participants can submit preliminary results and receive feedback on relative scoring for unlimited number of times.
- April 25, 2025: Phase III starts. Final test datasets are made available.
- May 13, 2025: Deadline for testing phase.
- May 20, 2025: Participants are notified with Challenge results and winners (top 2) are announced.
- July 27-31, 2025: AAPM Annual Meeting & Exhibition: top two teams will present on their work during a dedicated Challenge session.
- September 2025: The Challenge organizers summarize the Grand Challenge in a journal paper. Training and validation datasets and scoring routines will be made public.
Results, Prizes and Publication Plan:
Monetary awards. Total $4,000 for top 5 teams (pending official approval).
Authorship. (1) For the top 5 teams, we invite up to two members of each team as co-authors of the manuscript summarizing the Challenge results and submit the manuscript to a high-impact journal. Additional members of top teams will be included in the acknowledgement. (2) Top 2 solutions are invited to submit a manuscript to Medical Physics.
AAPM Annual Meeting. The top 2 teams (one member per team) will be invited to the AAPM Annual Meeting to present on their results during a dedicated Challenge session.
Internship opportunities. For the top 3 teams, if the first author of the team is a full-time student, we offer research internship opportunities at the AI Center of Siemens Healthineers in Princeton NJ (with priority, where only the last round of interviews would be required).
Terms and Conditions:
- Anonymous participation is not allowed and only one representative can register per team.
- No extra data / annotation is allowed.
- Members from the host institution and the organizers’ groups cannot participate. Please review and agree to AAPM's participant COI statement.
- Entry by commercial entities is permitted but should be disclosed; conflict of interest attestations will be required for all participants upon registration.
- Once participants submit their results to the GDP-HMM Challenge, they will be considered fully vested in the Challenge, so that their performance results will become part of any presentations, publications, or subsequent analyses derived from the Challenge at the discretion of the organizers.
- Participants summarize their algorithms in a document to submit at the end of Phase III. The participants need to submit their training code to ensure reproducibility to get awarded.
- Participants follow the citation requirement documented in https://github.com/RiqiangGao/GDP-HMM_AAPMChallenge.
Organizers:
- Riqiang Gao, Ph.D., lead organizer, (Siemens Healthineers)
- Florin Ghesu, Ph.D., (Siemens Healthineers)
- Wilko Verbakel, Ph.D., (Varian, a Simens Healthineers company)
- Rafe Mcbeth, Ph.D., (University of Pennsylvania)
- Sandra Meyers, Ph.D., (UC San Diego Health)
- Masoud Zarepisheh, Ph.D., (Memorial Sloan Kettering Cancer Center)
- Ali Kamen, Ph.D., (Siemens Healthineers)
- AAPM’s Working Group on Grand Challenges
Contacts:
For further information, please contact the lead organizer, Riqiang Gao (riqiang.gao@siemens-healthineers.com) or AAPM staff member, Emily Townley (emily@aapm.org).
References:
- [1] Wang et al. Deep Learning-Based Head and Neck Radiotherapy Planning Dose Prediction via Beam-Wise Dose Decomposition. MICCAI 2020.
- [2] Gao et al. Flexible-Cm GAN: Towards Precise 3D Dose Prediction in Radiotherapy, CVPR 2023.
- [3] Babier et al. OpenKBP: The Open-Access Knowledge-Based Planning Grand Challenge. Medical Physics, 2020.
Disclaimer:
The resources and information provided in this Challenge are based on research results and for research purposes only. Future commercial availability cannot be guaranteed.



















