An AAPM Grand Challenge
Overview
The American Association of Physicists in Medicine (AAPM) is sponsoring the DGM-Image Challenge – a Grand Challenge on deep generative modeling for learning medical image statistics, leading up to the 2023 AAPM Annual Meeting. This Challenge invites participants to develop or refine generative models that can accurately reproduce image statistics important and relevant to medical imaging applications. An individual from each of the two top-performing teams will be invited to present their results during the AAPM 2023 Annual Meeting Grand Challenge Symposium and will be awarded a complimentary meeting registration to attend the meeting. The findings and insights from the Challenge will be published via a Challenge report.
Background
Over the past few years, deep generative models, such as generative adversarial networks (GANs) and diffusion models, have gained immense popularity for their ability to generate perceptually realistic images [1,2,3]. Deep generative models (DGMs) are being actively explored in the context of medical imaging applications, such as data sharing, image restoration, reconstruction, translation, and objective image quality assessment [4,5]. State-of-the-art DGMs trained on medical image datasets have been shown to produce images that look highly realistic, but nevertheless may contain medically impactful errors [4,6]. Unlike medical image classification and, more recently, medical image reconstruction, there is a glaring lack of standardized evaluation pipelines and benchmarks for developing and assessing deep DGMs for medical image synthesis. This Challenge aims to establish DGMs that can faithfully reproduce image statistics relevant to medical imaging. Such models, when validated, can play a key role in the evaluation of imaging systems as well as for use in training and/or testing of AI/ML algorithms, especially for applications where clinical data is limited.
Objective
The goal of this Challenge is to facilitate the development and refinement of generative models that can reproduce several key image statistics that are known to be useful for a wide variety of medical image assessments. Though this Challenge, a custom image dataset of coronal slices from anthropomorphic breast phantoms based on the VICTRE toolchain [7] will be provided to the participants to train their generative models. This Challenge will identify the generative model learned from the provided data that most accurately reproduces the training data distribution of morphological and intensity-derived statistical measures as well as breast-density relevant features, while still producing perceptually realistic images and avoiding overfitting/memorization of the training data. An eventual goal of this Challenge is to provide a dataset, a standardized evaluation procedure, and a benchmark for evaluating generative models for medical image synthesis.
This Challenge will be divided into two phases. Phase 1 will require the participants to submit a dataset of 10,000 images from their generative model along with a 1-page description of their approach. These will be used to compute performance measures and populate the leaderboard. Phase 2 of the Challenge will require the participants to submit their code for generating an image dataset. In Phase 2, code submission from at least top 10 submissions will be manually checked and images will be re-generated using the participant code to verify the results.
Get Started
- Register to get access via the Challenge website.
- Download the data (and code) after approval.
- Train or develop your generative model.
- (Phase 1) Submit generated images and description of approach to the Challenge website.
- (Phase 2) Submit generated images, code, and description of approach to the Challenge website.
Important Dates
- January 13, 2023: Challenge announced, participant registration opens.
- January 16, 2023: Training set release
- May 19, 2023: Deadline for Phase 1 submission
- May 26, 2023: Deadline for Phase 2 submission
- June 16, 2023: Participants and top-ranked finishers contacted with results
Challenge Methodology
- Dataset:
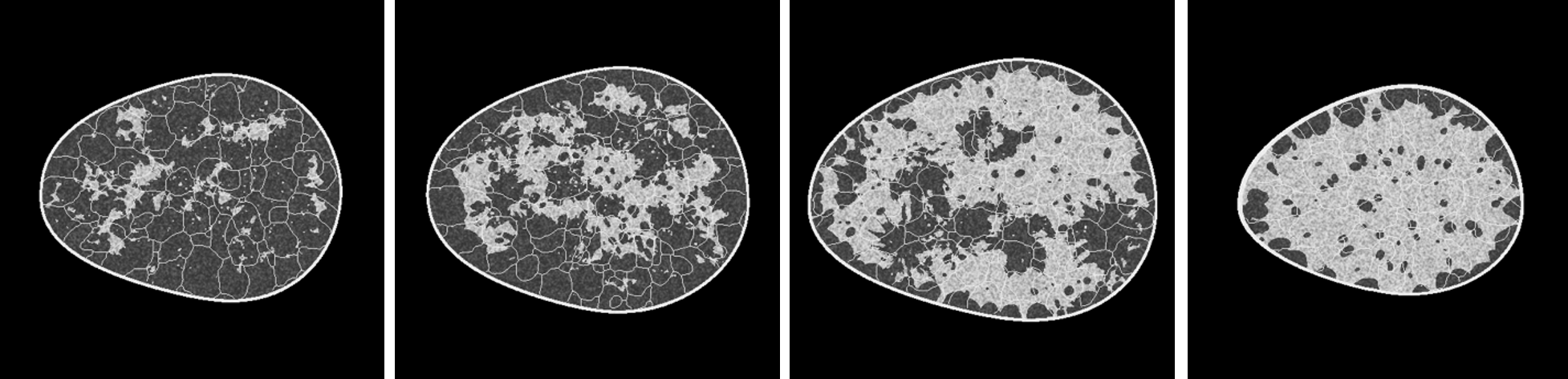
The provided training dataset is an unlabeled dataset containing 100,000 8-bit images of size 512x512. Based off the VICTRE breast phantom creation software, these images emulate coronal slices from anthropomorphic breast phantoms. They contain fibroglandular structures interspersed with fatty tissue and ligament networks. Appropriate X-ray attenuation coefficients are assigned to the various tissues based on existing breast computed tomography (bCT) literature, along with added texture. The slices are then rescaled and digitized to 8-bit images. The data roughly comprises four breast types – fatty, scattered, heterogeneous and dense – based on the Breast Imaging Reporting and Data System (BI-RADS) classification of breast density [8].
- Evaluation metrics:
Our evaluation pipeline will be divided into two stages:
Stage I: In stage I, the submitted models will be evaluated using well-known perceptual measures such as the Frechet Inception Distance (FID) score. A custom memorization metric will also be used. Models (such as Memory-GANs) that display memorization of the training dataset or fail to produce perceptually realistic images in terms of the FID score will be disqualified based on a suitable threshold. The models that qualify in this stage will be further evaluated via Stage II.
Required Items for Phase 1 Submissions
- 10,000 images generated from your model.
- A 1-page description of your approach.
- Intensity-derived statistics such as gray-level texture features,
- Morphological statistics, such as region properties, skeleton statistics, and fractal dimension,
- Breast-density relevant features.
- 10000 images generated from your model.
- Code, trained model weights, and a bash script to run the model to generate 10,000 images along with a well-tested dockerfile that containerizes your code
- A 1-page description of your approach. This must contain a time estimate for generating 10,000 images on a single Nvidia V100 GPU.
To assist the participants in developing their best models, we provide a basic implementation of the stage II of our evaluation (final evaluation may be slightly different). We also provide the scores for a baseline model (StyleGAN2) along with the trained weights, which the participants are allowed to use for initialization/transfer learning.
Terms and Conditions
- Participants must ensure that their model can generate 10000 images within 12 hours on a single V100 GPU. If it takes >12 hours to generate 10000 images, the model may be disqualified depending upon our computational budget.
- At the end of the Challenge, code submission from at least top 10 submissions will be manually checked and images will be re-generated using the participant code to ensure that there is no cheating or memorization.
- Participants will share the names of their team members and contact information during registration. However, this will not be made public on the leaderboard.
- Participants are only allowed to use the provided training data. Participants are allowed to use publicly available weights of pretrained models for the purpose of transfer learning, as long as the details of what weights are used is provided in the 1-page description during submission.
- Participants are not allowed to submit images from the training data or other forms of memory models. Submissions are restricted to machine learning models that are estimated from the provided training data. This will be checked during the code review at the end of the competition.
- Descriptions of participants’ methods and results may become part of presentations, publications, and subsequent analyses derived from the Challenge (with proper attribution to the participants) at the discretion of the Organizers. While methods and results may become part of Challenge reports and publications, participants may choose not to disclose their identity and remain anonymous for the purpose of these reports.
- Please note, one member from each top-ranked team will be awarded complimentary meeting registration to attend AAPM's 2023 Annual Meeting in Houston, TX (July 23-27, 2023), in-person attendance is mandatory. They will present on their results and methodology during a Grand Challenge Symposium and participate in an Awards & Honors Ceremony.
Organizers (arranged alphabetically)
Mark A. Anastasio1, Frank J. Brooks1, Rucha M. Deshpande2, Dimitrios S. Gotsis1, Varun A. Kelkar1, Prabhat K.C.3, Kyle J. Myers4, Rongping Zeng3.
1 University of Illinois at Urbana-Champaign
2 Washington University in St. Louis
3 US Food and Drug Administration
4 Puente Solutions, LLC
AAPM's Working Group on Grand Challenges
Contact: dgm.image@gmail.com
References
- Foster, David. Generative deep learning: teaching machines to paint, write, compose, and play. O'Reilly Media, 2019.
- Karras, Tero, et al. "Analyzing and improving the image quality of stylegan." Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2020.
- Dhariwal, Prafulla, and Alexander Nichol. "Diffusion models beat gans on image synthesis." Advances in Neural Information Processing Systems 34 (2021): 8780-8794.
- DuMont Schütte, August, et al. "Overcoming barriers to data sharing with medical image generation: a comprehensive evaluation." NPJ digital medicine 4.1 (2021): 1-14.
- Yi, Xin, Ekta Walia, and Paul Babyn. "Generative adversarial network in medical imaging: A review." Medical image analysis 58 (2019): 101552.
- Kelkar, Varun A., et al. "Assessing the ability of generative adversarial networks to learn canonical medical image statistics." arXiv preprint arXiv:2204.12007 (2022).
- Badano, Aldo, et al. "Evaluation of digital breast tomosynthesis as replacement of full-field digital mammography using an in-silico imaging trial." JAMA network open 1.7 (2018): e185474-e185474.
- D’Orsi, Carl, L. Bassett, and S. Feig. "Breast imaging reporting and data system (BI-RADS)." Breast imaging atlas, 4th edn. American College of Radiology, Reston (2018).



















